PseCas9
A sticky-cutter enzyme from Cas9-IIBs
Background - CRISPR-Cas in bacteria
CRISPR-Cas9 was originally adapted from bacterial systems for precise genome editing in human cells. The discovery of CRISPR-Cas9 systems was driven by the pioneering work of Prof. Jennifer Doudna and Prof. Emmanuelle Charpentier, who sought to understand how bacteria acquire immunity against phages.
In prokaryotes, CRISPR-Cas systems are multi-component complexes comprising one or more Cas proteins (e.g., Cas1, Cas2) and short RNAs (e.g., crRNA and tracrRNA). Together, these components form a RiboNucleoProtein (RNP) complex. In most bacteria, multiple Cas proteins collaborate to identify and destroy the genetic material of invading viruses. However, in a small subset of bacteria, a single protein, Cas9, performs all the necessary functions to locate and eliminate the invading genomes.
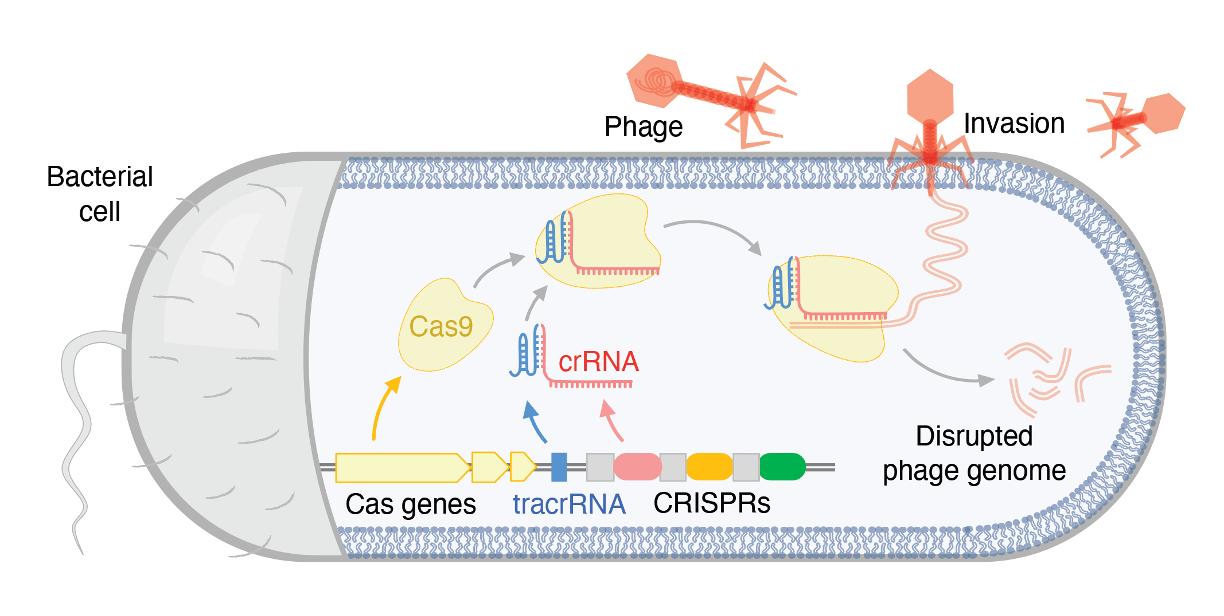
The challenge - harnessing novel CRISPR-Cas systems
To date, the most widely used CRISPR-Cas9 system in human cells is the Cas9 protein discovered in Streptococcus pyogenes (SpyCas9). Adapting new CRISPR-Cas9 systems from prokaryotic cells for use in human cells presents several challenges.
First, correctly identifying RNA species (e.g., crRNA and tracrRNA) from raw, unannotated sequencing data requires multidisciplinary expertise in bioinformatics and molecular biology. Second, fundamental differences in the central dogma of life between prokaryotic and eukaryotic cells add another layer of complexity, particularly when testing the activity of new Cas9 variants in human cells.
The acheivement
During my postdoctoral research at AstraZeneca, I contributed to the discovery and adaptation of several Cas9 proteins from the IIB class. We found that Cas9 proteins from this class cut DNA at different positions compared to the commonly used SpyCas9.
While spCas9 generates blunt ends or single-nucleotide overhangs during DNA cleavage, IIB-class Cas9s cleave DNA in a staggered manner, leaving 3- to 5-nucleotide overhangs (Bestas et al., 2023). This novel nuclease activity has the potential to facilitate the development of more efficient knock-in strategies.
For more details, see patent number: WO2019099943A1
References
2023
-
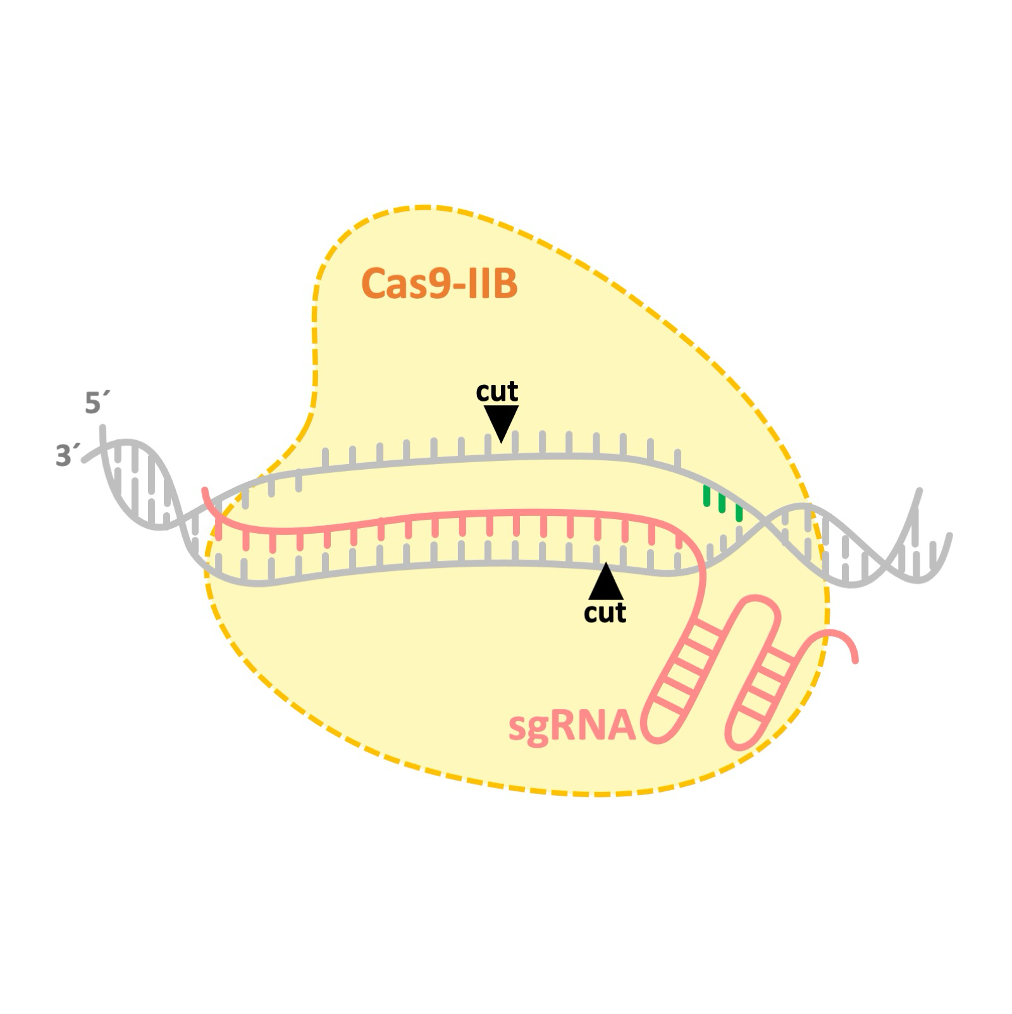 A Type II-B Cas9 nuclease with minimized off-targets and reduced chromosomal translocations in vivoNature Communications, 2023
A Type II-B Cas9 nuclease with minimized off-targets and reduced chromosomal translocations in vivoNature Communications, 2023
2022
-
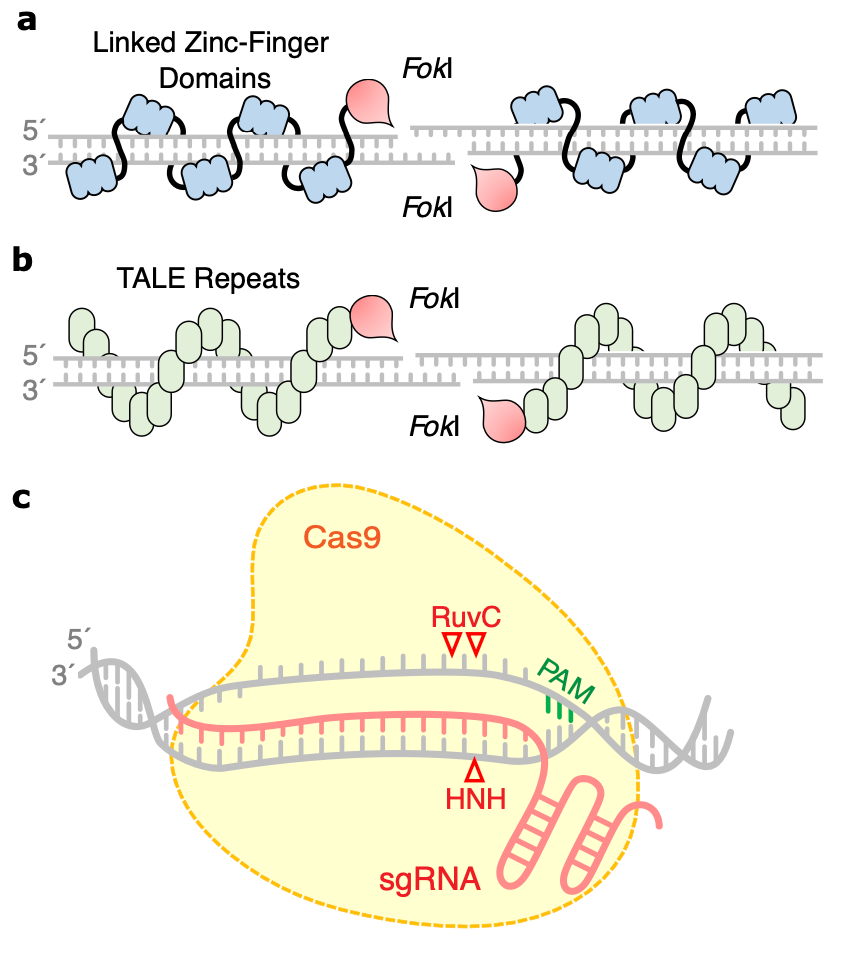 Tools for efficient genome editing; ZFN, TALEN, and CRISPRApplications of genome modulation and editing, 2022
Tools for efficient genome editing; ZFN, TALEN, and CRISPRApplications of genome modulation and editing, 2022