RIMA
Rational Indel Meta-Analysis
Background - “Random Indels”
Genome editing involves two sequential steps: cutting and repairing. The cut is typically introduced by programmable nucleases, such as Cas9, in a targeted manner. The repair process, however, primarily depends on the cell’s own DNA repair mechanisms. If repair perfectly restores the sequence at the cleavage site, the targeted site may be cleaved repeatedly by the nuclease. However, repairs can also introduce new mutations, such as insertions and deletions (commonly referred to as indels), at the cleavage site.
The Challenge - DNA Repair Profiling
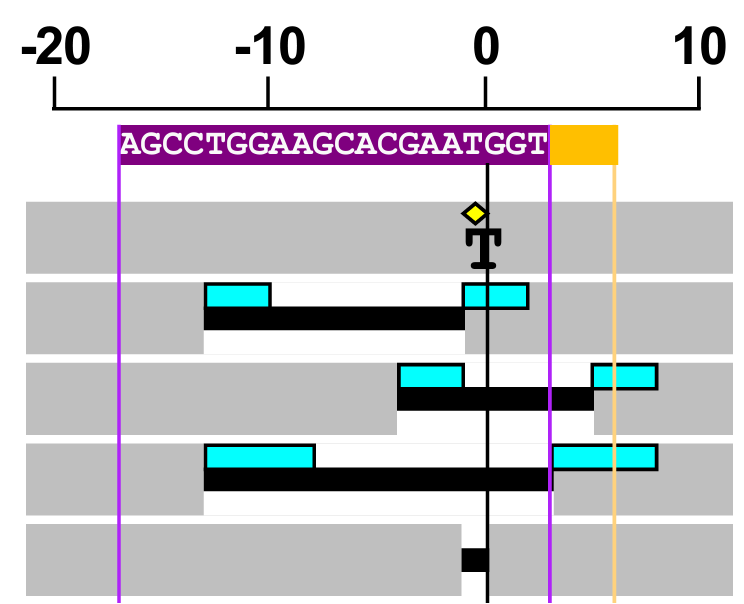
Indels were long assumed to be random repair events. However, recent studies have demonstrated their non-random nature. Indel profiles at different genomic loci are distinct, yet these profiles are reproducible at the same target site across different cell lines and experimental replicates. This raises an intriguing question: What drives the formation of reproducible mutations at specific target sites in different cells?
The Achievement - Rational Indel Meta-Analysis (RIMA)
During my research on characterizing Cas9 nucleases in human cells, I developed a bioinformatics tool called Rational Indel Meta-Analysis (RIMA) (Taheri-Ghahfarokhi et al., 2018). RIMA analyzes, categorizes, and visualizes mutation patterns at Cas9 cleavage sites, shedding light on how DNA repair pathways interact with Cas9 activity and the sequence context of target sites. This tool helps explain the mechanisms underlying the non-random nature of Cas9-induced mutations.
RIMA also profiles the contribution of different DNA repair pathways in the repair of DNA double-strand breaks. It has facilitated various applications in genome editing and cancer research. For example, I used RIMA to distinguish the nuclease activity of spCas9 and new Cas9 proteins from the IIB class, demonstrating how different Cas9 variants produce distinct indel profiles at their target sites.
In cancer research, cells often lose the function of one DNA repair pathway, forcing them to rely on the remaining pathways to support their rapid replication. This dependency makes overactive DNA repair pathways promising targets for cancer drug discovery. Using RIMA, we can measure the overall DNA repair capacity of cells, validate the roles of key proteins in DNA repair pathways, and identify compounds that inhibit overactive repair mechanisms.
For further details, see my publication in Nucleic Acids Research here.
References
2018
-
 Decoding non-random mutational signatures at Cas9 targeted sitesNucleic acids research, 2018
Decoding non-random mutational signatures at Cas9 targeted sitesNucleic acids research, 2018